Research

Our goal:
What distinguishes a living system from a non-living system? Both are made of atoms and governed by the same physical laws. Neither one atom nor two atoms forms a living system. But step by step, as we connect three, four, and eventually many Avogadro numbers of atoms together in just the "right" way, we obtain a complex system characterized by dynamics that we associate with life - a living system. Indeed, living systems are dynamic. We associate motion, chemical reactions, and information processing with being alive. But non-living systems can also be dynamic. How do the dynamics of living matter differ from those of non-living matter?
Long-lived, complex dynamics of living systems:
"Living matter evades the decay to equilibrium", according to Schrodinger. That is, living matter generates and maintains systems-level dynamics - involving many components and processes - that do not halt for a long time, lasting longer than the lifetimes of the molecules involved and beyond the timescales of the constituent chemical processes. These systems-level dynamics tend to be complex, exhibited by one or more cells.Our lab seeks to quantitatively explain the *capacity* that living systems have for generating and maintaining long-lived, complex dynamics that non-living matter hardly ever exhibits . Permanently losing that capacity, such as in dormancy, would mean that the cell or a multicellular system can no longer achieve those dynamics. Moreover, even if a system is not dynamic now, having the capacity to achieve those dynamics means that it can still restart and maintain long-lived, complex dynamics that we associate with life.
To achieve our goal, we perform real and computational experiments that we interpret through the language of dynamical systems. Thus far, we have been focused on two related, long-lived dynamics that are arguably unique to living systems: 1.) dynamics of self-replication and how a non-replicating cell maintains self-replicability; and 2.) self-organization.
1. Self-replication dynamics : limits to keeping it going and when it can stop and resume
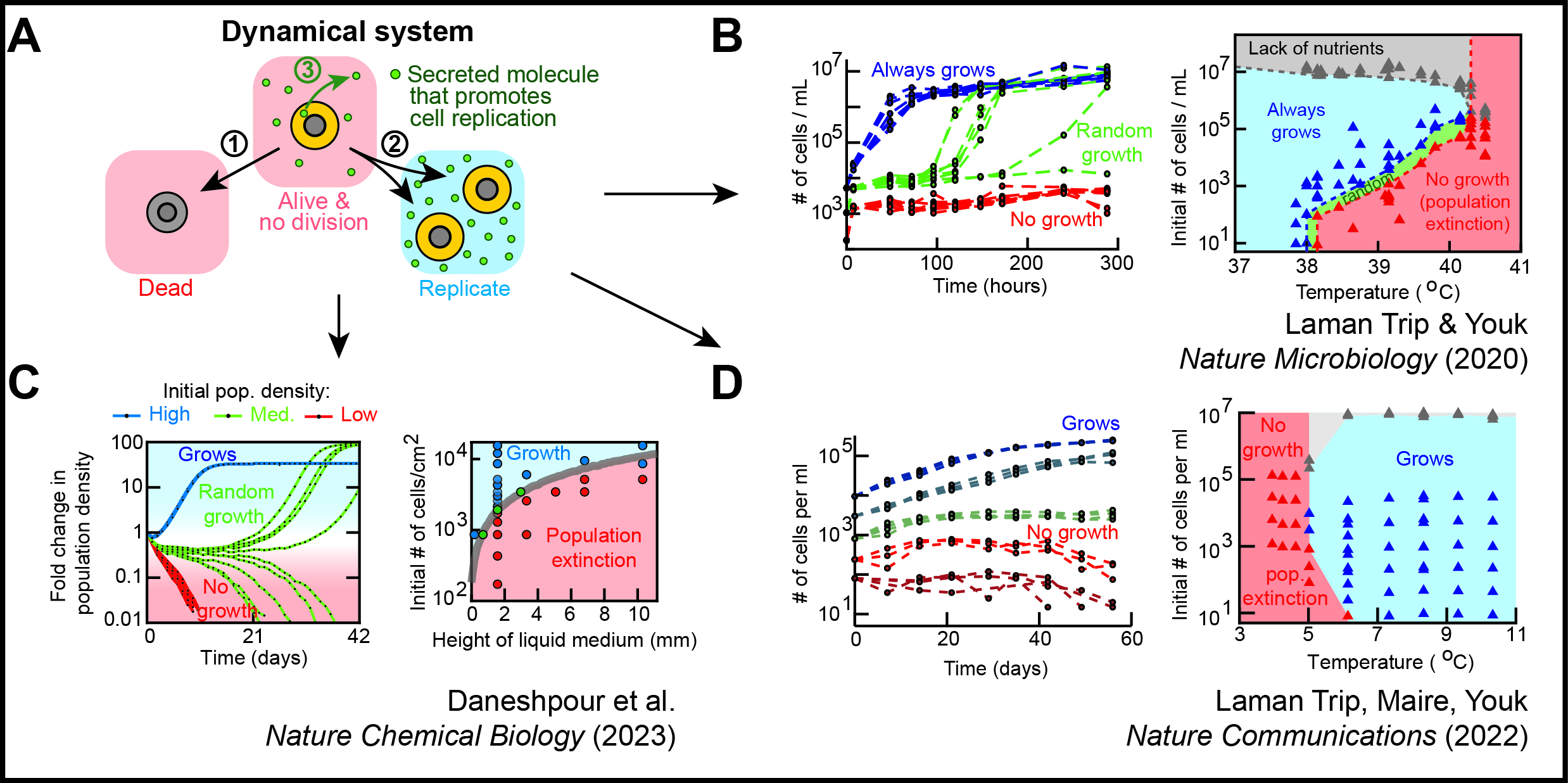
Cells maintain viability - their ability to self-replicate - for much longer than the lifetimes of their constituent molecules and the timescales of the chemical processes that coalesce to yield self-replication. Moreover, self-replication is a complex dynamics in that it appears to be functionally periodic - the cell repeatedly self-replicates - but is physically aperiodic because the cell does not have the same copy number of every molecule, each at exactly the same location, as it self-replicates again. Cells may also affect each other's replication, raising the complexity. As a dynamical system, how does a cell maintain its viability - its non-periodic self-replication dynamics - for a long time? Are there unbeatable constraints to maintaining this dynamics? To address these questions, we are using two approaches: (1) slowing down a cell's life to discover constraints to maintaing self-replication dynamics and (2) halting a cell's life and then resuming its life - examining "lifeless-to-alive" transition - to discover how a static cell can become dynamic (self-replicating) again.
Approach 1 - Slowing life to reveal limits of self-replication dynamics:
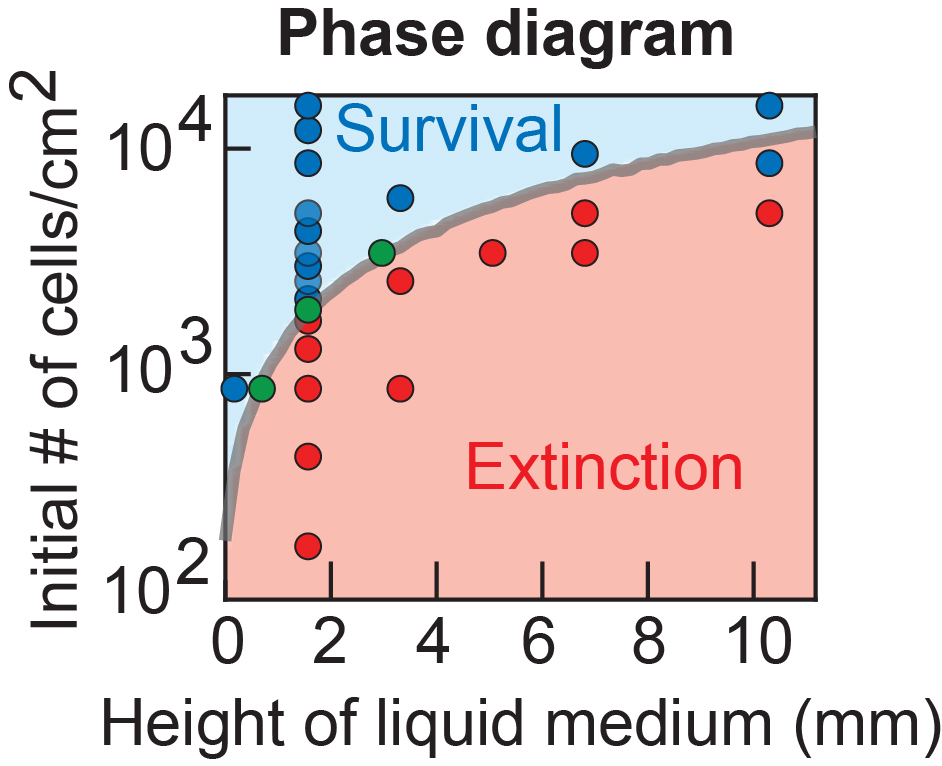
As we gradually slow down all intracellular processes, does a living system break down (stop living) at some point? Or can life progress at an arbitrarily slow speed? How do we even measure a "speed of life"? We're addressing these questions by drastically slowing a cell's self-replication dynamics by, for example, increasing or decreasing temperature to extreme values or inducing growth-slowing genetic programs (e.g., cell differentiation).
 |
H. Daneshpour, P. van den Bersselaar, C.-H. Chao, T. G. Fazzio, and H. Youk Macroscopic quorum sensing sustains differentiating embryonic stem cells Nature Chemical Biology (Jan. 2023) Online version (.html) News & Views on this work (.pdf) |
 |
D. S. Laman Trip, T. Maire, and H. Youk Slowest possible replicative life at frigid temperatures for yeast Nature Communications (Dec. 2022) Online version (.html) Article in PDF Supplementary Info + Theory (.pdf) Supp. Movies (.html) Twitter Tweets summarizing this work |
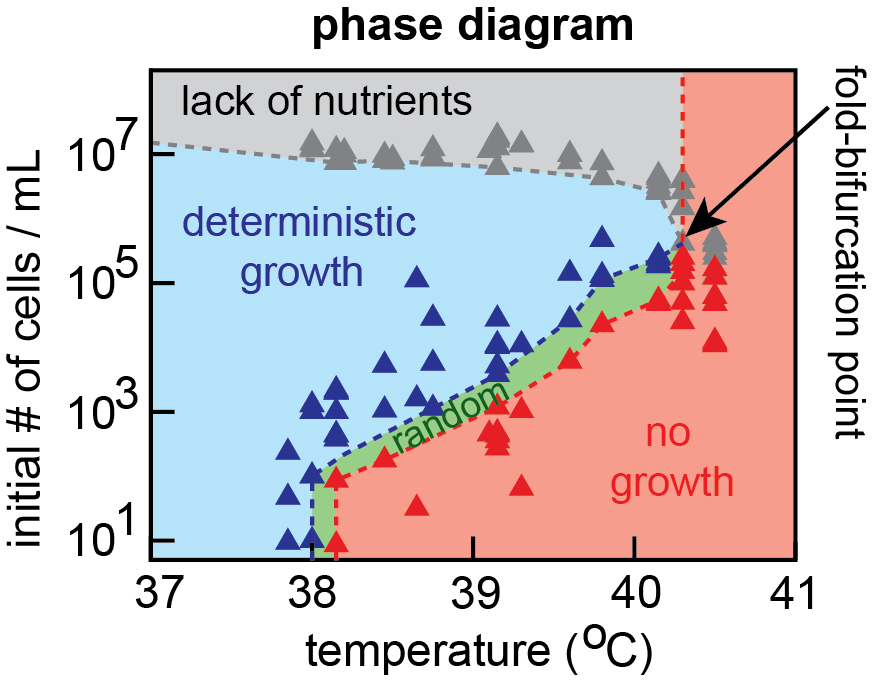 |
D. S. Laman Trip and H. Youk Yeasts collectively extend the limits of habitable temperatures by secreting glutathione Nature Microbiology (April 2020) Online version (.html) Article in PDF News & Views (.pdf) Twitter Tweets summarizing this work |
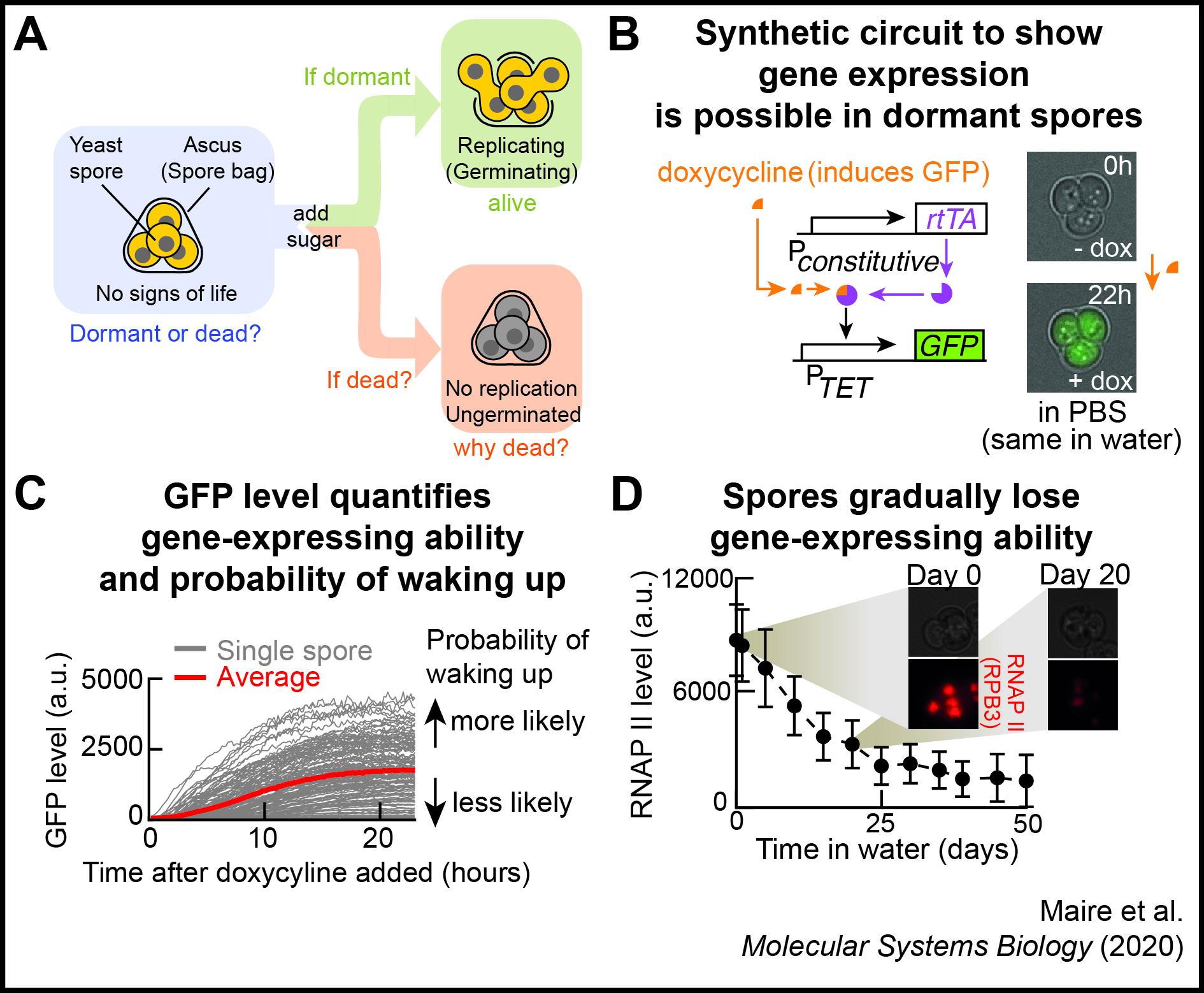
Approach 2 - Restarting halted life to understand lifeless-to-alive transition
Can we completely stop any living system and then resume its life by presssing a "button" as in sci-fi movies? If yes, then how? If not, then why can't the dynamical system (cell) restart its self-replicative dynamics? We are investigating this question with orgnanisms whose lives have halted and thus seem lifeless while maintaining the ability to resume self-replicatiion (e.g., dormant yeast spores).
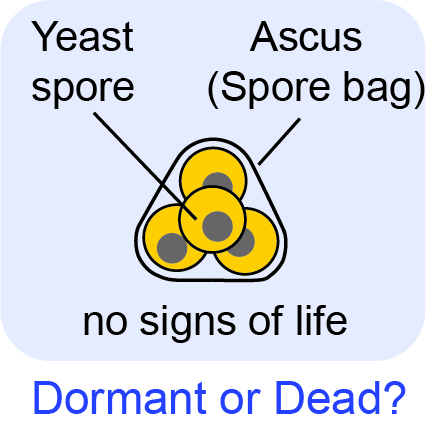 |
T. Maire, T. Allertz, M. A. Betjes, and H. Youk Dormancy-to-death transition in yeast spores occurs due to gradual loss of gene-expressing ability Molecular Systems Biology (November 2020) Online version (.html) Article in PDF Twitter Tweets summarizing this work |
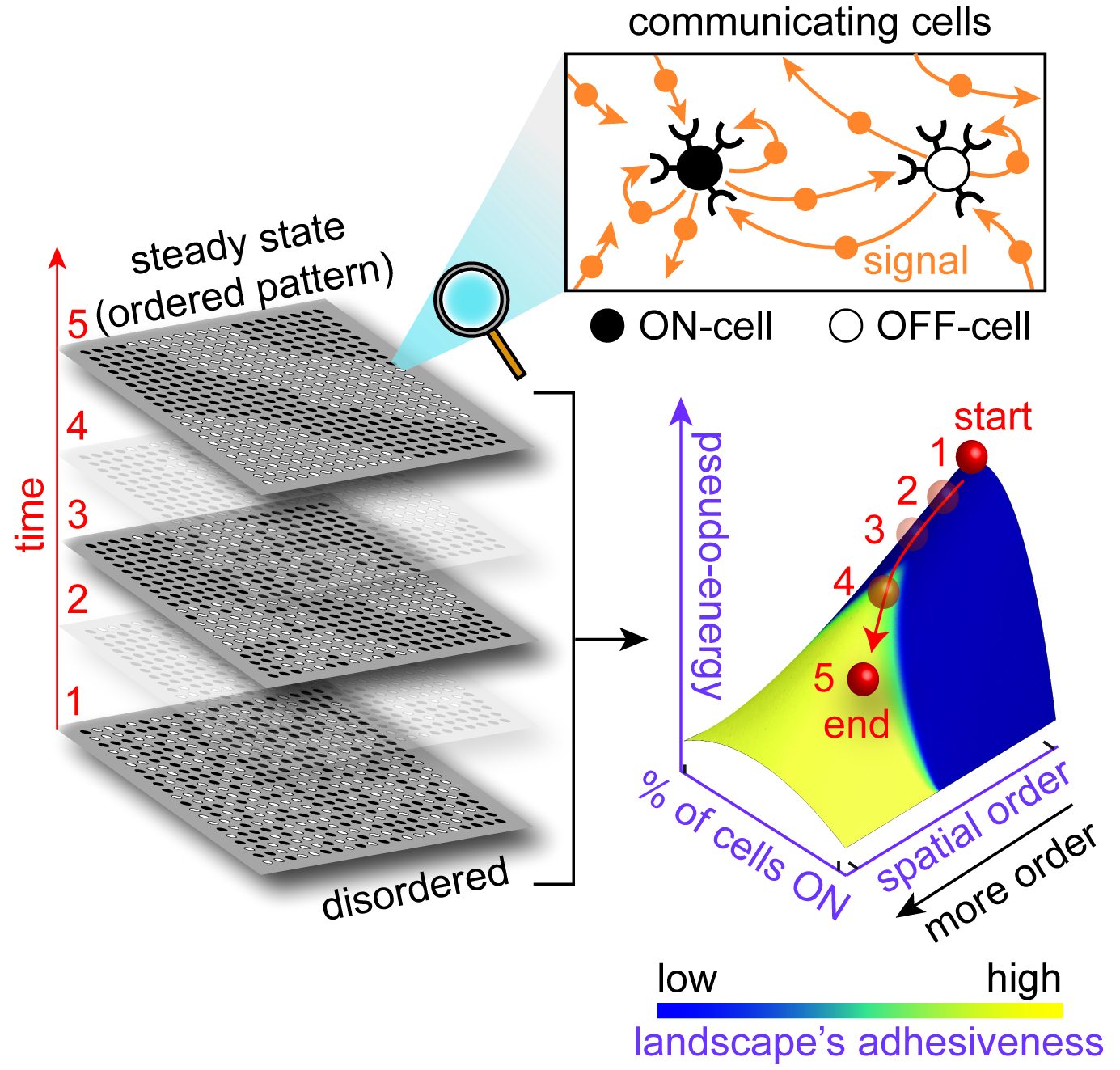
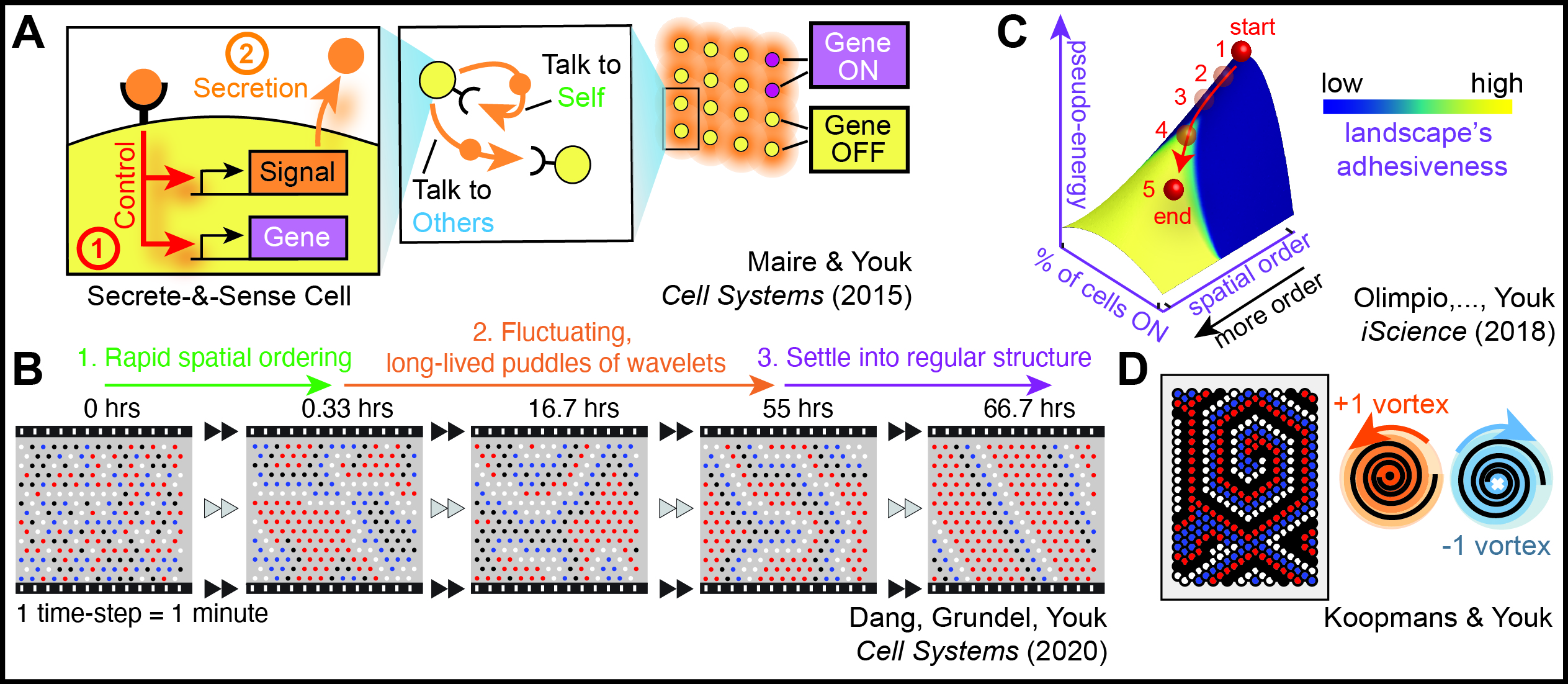
2. Self-organization dynamics : Computational experiments to reveal dynamics of spatial-order generation by cells
Living systems excel at forming ordered structures from disorganized components, as exemplified by cell replication. To understand self-organization dynamics, particularly spatial pattern formation, we use cellular automata as computational experiments. These automata employ discrete approximations, such as binary actions, to represent biological processes. Binary actions like "protein A is either bound or not bound to protein B" and "gene is either highly or lowly expressed" can effectively capture essential cellular rules. Incorporating non-nearest-neighbor interactions in our cellular automata helps reveal large-scale and long-lasting dynamics characteristic of spatial-pattern-forming cells.
We analyze known and hypothetical gene regulatory rules using cellular automata to identify those that enable cells to form spatially ordered configurations, such as spiral waves. Additionally, we develop methods to quantify the evolving spatial order in a field of communicating cells forming patterns. While certain spatial patterning mechanisms are well-understood, our knowledge of pattern formation through non-local interactions, like diffusing molecules used for gene regulation, remains limited.
Our research shows that non-locally interacting cells can generate complex, spatially organized dynamics, some of which are observed in nature, while others await discovery. Our objective is to investigate all potential cellular dynamics generated by biologically relevant discrete rules using cellular automata, offering a comprehensive perspective on living systems as discrete, dynamical systems and differentiating them from non-living systems.
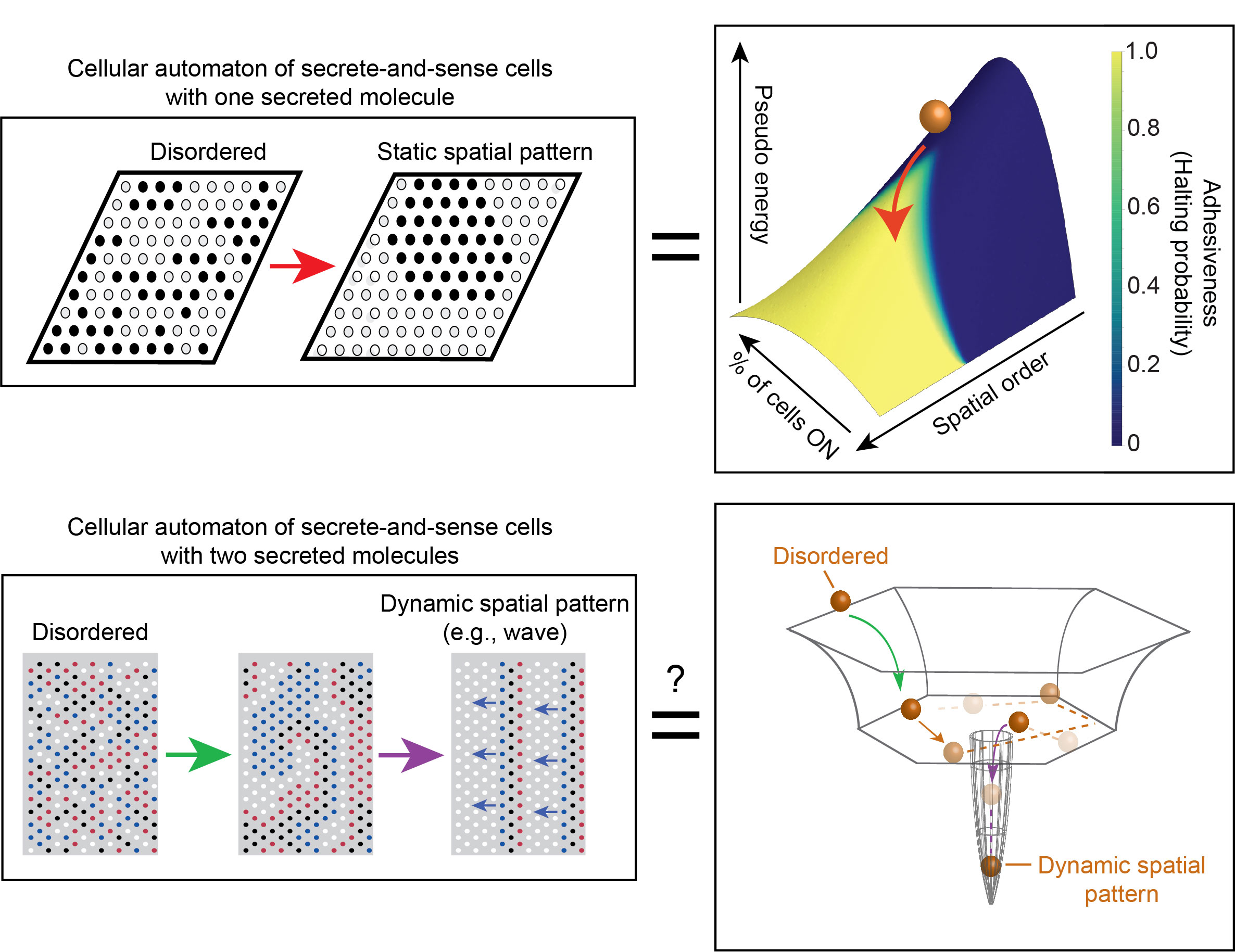 |
L. Koopmans and H. Youk Predictive landscapes hidden beneath biological cellular automata Journal of Biological Physics (Nov. 2021) Online version (.html) review (.pdf) |
 |
Y. Dang, D. A. J. Grundel and H. Youk Cellular dialogues: cell-cell communication through diffusible molecules yields dynamic spatial patterns Cell Systems (January 2020) Online version (.html) Article in PDF |
 |
E. P. Olimpio*, Y. Dang*, and H. Youk (*Co-first authors) Statistical dynamics of spatial-order formation by communicating cells iScience (April 2018) Online version (.html) Article + Supp. Info (.pdf) |

|
T. Maire and H. Youk |





 Click on the image to learn more.
Click on the image to learn more.
 Click on the image to learn more.
Click on the image to learn more.
 Click on the image to learn more.
Click on the image to learn more.